Seshat:
 |
Seshat:* An innovative tool to handle somatic and germline TP53 variants.
Users can test a single variant on this page or perform a batch analysis using txt file as well as VCF or MAF file generated by NGS. Seshat performs the following tasks: Seshat works with both csv files and vcf or maf files generated by NGS. Input data using protein, cDNA or genomic information can be used. Seshat is based on the UMD TP53 database; 2017 release: 80,000 mutations (6,000 TP53 variants).
* Seshat was the Ancient Egyptian goddess of wisdom, knowledge, and writing. Her name means she who is the scribe, which is in perfect harmony with the goal of this site.* |
Seshat: Where ?
You can access Seshat via this link: beware that you will leave the TP53 web site to access Seshat
Seshat: Why ?
Somatic mutations in the TP53 gene are one of the most frequent alterations in human cancers, and germline mutations are the underlying cause of Li-Fraumeni syndrome, which predisposes to a wide spectrum of early-onset cancers.
Accurate assessment of TP53 gene status in sporadic tumors and in the germline of individuals at high risk of cancer has important clinical implications for diagnosis, surveillance and therapy.
The most recent version of the National Comprehensive Cancer Network (NCCN) guidelines recommends TP53 mutations testing in individuals with onset of breast cancer before 31 years of age, either concurrently with BRCA1/2 testing or as a follow-up test after negative BRCA1/2 testing
(NCCN Guidelines Version 1.2017, https://www.nccn.org/professionals/)
Somatic TP53 mutation analysis is now widely used in clinical trials involving patient stratification based on TP53 status and in trials of novel drugs targeting either wild-type or mutant TP53 in order to activate a TP53 antitumor response. TP53 mutation screening is therefore rapidly becoming an integral part of many therapeutic or prevention strategies in clinical practice.
The complex architecture and expression pattern of the TP53 gene has only been recognized in recent years. TP53 mobilizes various mechanisms to transcribe at least eight different mRNA isoforms, which are generated by alternative splicing or alternative promoter usage. Collectively, these mRNAs have the potential to give rise to up to 12 different proteins, although the exact expression level, tissue distribution and biological function of each of these protein variants are poorly understood. This complex expression pattern implies that sequences located in TP53 introns and involved in the production of alternative forms of the protein may have a critical impact on overall biological functions of p53, and may therefore be important target regions for somatic or germline variants.
Bioinformatic pipelines currently associated with various NGS devices use different nomenclatures, algorithms and references to assess TP53 status, leading to heterogeneous outputs. For example, we have observed that variant chr17:g.7578406G>A (MN_000546.5_c.524G>A) can be randomly described as either p.R175H (using NP_000537.3 as a reference) or p.R43H (using NP_001119587.1 as a reference) in the same output. The fact that no reference is usually included results in the misleading conclusion that two different TP53 variants have been identified.
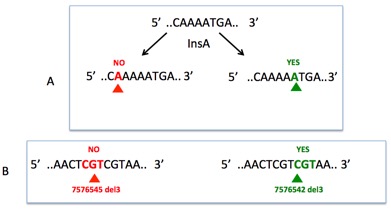
Another problem concerns the mutation nomenclature, which is not homogeneous among the various outputs and rarely follows the hgvs recommendations (http://varnomen.hgvs.org/), leading to the false impression that a given mutation is a novel mutation (Figure ). The 3' rule used for the description of mutations in repeated sequences is rarely followed.
|
The 3' rule, a misapplied rule: For all descriptions, the most 3' position possible of the reference sequence is arbitrarily assigned to have been changed. A: Insertion of nucleotide A indifferently at various positions leads to multiple mutational events despite a similar outcome. B: Deletion of multiple nucleotides can lead to the same final sequence. The most 3' event leading to the final variant must be used to generate a unique and reproducible mutational event. Seshat was developed to resolve all of these problems. Using raw data (vcf, maf or txt files), Seshat generates multiple output tables with accurate and complete information on each TP53 variant.
|
More info here:
http://varnomen.hgvs.org/recommendations/general/